We present here the work we pursue in the Laboratoire Jean Perrin (CNRS - UPMC) ie. developing an "artificial morphogenesis" through DNA-based molecular programming and kinesin/microtubule active matter. We join the lab in March 2015. Previously we were working in the Laboratoire de Photonique et de Nanostructures, now Center for Nanosciences and Nanotechnology.
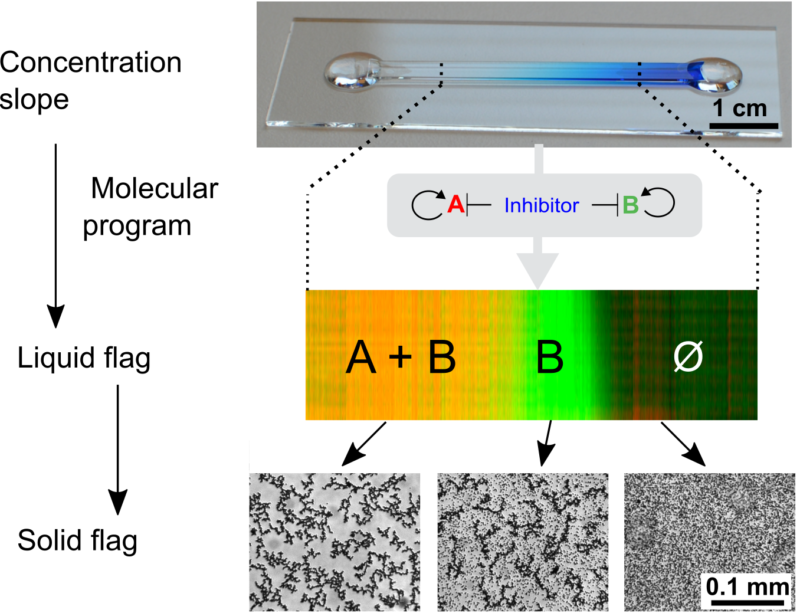
Figure 1: A "life-like" material through reaction-diffusion patterning. A molecular program based on short DNA strands and enzymes is able to read and interpret a pre-existing gradient of immobile DNA to generate several chemically distinct zones. As for cells in an embryo, the fate of DNA-coated beads (translated into a level of aggregation) depends on the position. More information here.
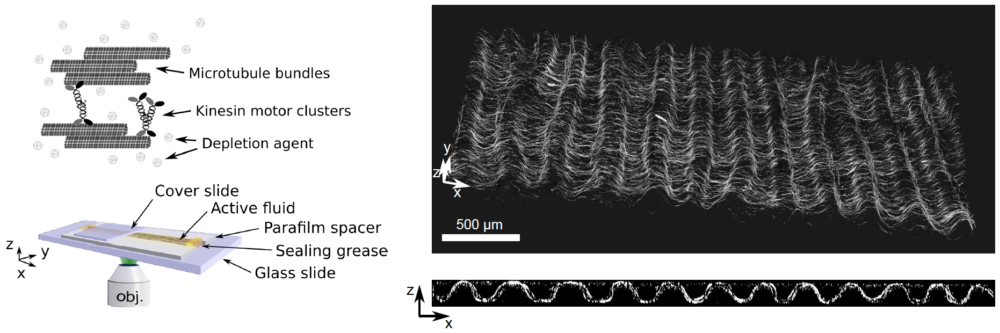
Figure 2: Active matter locally converts chemical energy into mechanical work and, for this reason, it provides new mechanisms of pattern formation. In particular, active nematic fluids made of protein motors and filaments are far-from-equilibrium systems that may exhibit spontaneous motion. We demonstrated that a 3D solution of kinesin motors and microtubule filaments can also spontaneously form a 2D free-standing nematic active sheet that actively buckles out-of-plane into a centimeter-sized periodic corrugated sheet that is stable for days. More information here.

Figure 3: A combination of self-organizating mechanisms. Here, we conjugate the two out-of-equilibrium patterning mechanisms we use - reaction-diffusion and active matter - to mimick key aspects of the polarization mechanism observed in C. elegans oocytes. A mechano-chemical transduction is produced: the microtubules-based active matter contracts globally, triggering a DNA-based reaction-diffusion front. More information here.
News
- We are looking for motivated students to join our group for an internship (M1/M2).
- Apr. 2025. Publication in PRX
- Aug. 2024. Baptiste is leaving. We wish you the best in Nice!
- Apr. 2024. Gwenael is leaving. We wish you the best at Essilor!
- Jan. 2024. Publication in PNAS
- Oct. 2023. A new post doc in the lab: Gwenael Bonfante. Welcome.
- Sept. 2023. Guillaume is leaving. We wish you all the best for the future!
- June. 2023. Guillaume is now a Doctor... Congratulations!
- June. 2023. Nicolas's work has been selected for an oral presentation at DNA29 conference (Tohoku University, Sendai). Congratulations!
- Feb. 2023. A new PhD student in the lab : Romain Leroux. Welcome.
- Feb. 2023. A new post doc in the lab: Baptiste Blanc. Welcome.
- July 2022. Good news: our ANR project LiliMat ("Life-Like" Materials, with F. Robin from Laboratoire de Biologie du Développement at Sorbonne Université) has been selected.
- April. 2022. Publication in Soft Matter
- April 2022. A new post doc in the lab: Nicolas Lobato-Dauzier. Welcome.
- Feb. 2022. Publication in ACS Synthetic Biology
- Jan. 2022. Publication in Physical Review E
- Jan. 2022. Marc is leaving. We wish you the best for the future!
- Dec. 2021. Publication in Science Advances
- Nov. 2021. Publication in JACS
- Sept. 2021. Yuliia is leaving. We wish you the best in Cambridge!
- Sept 2021. The MoleculArxiv project has been selected and will start next year. Together with Y. Rondelez (ESPCI), M. Delarue and M. Hollenstein (Pasteur Institute) and A. Genot (LIMMS Tokyo) we are part of the "DNA synthesis" axis of the MoleculArxiv project.
- Apr. 2021. A chapter published in a Wiley book
- Apr. 2021. Anis is leaving. We wish you the best for the future!
- Dec. 2020. Publication in ACS Nano
- Dec. 2020. A chapter published in a Wiley book
- Feb. 2020 Yuliia, post doc in the group, received funding from a Marie Sklodowska-Curie grant. Congratulations!
- Oct. 2019. Publication in Soft Matter
- Oct. 2019. Shunichi is leaving. We wish you the best in Japan!
- Oct. 2019. Publication in PNAS
- Sept. 2019. A new PhD student in the lab: Guillaume Sarfati. Welcome.
- May 2019. A new post doc in the lab: Yuliia Vyborna. Welcome.
- May 2019. Publication in ACS Biochemistry
- March 2019. After two years in the group, Georg is leaving. We wish you the best in Japan!
- Nov. 2018. Our new microscopy setup for cell culture is now fully installed.
- Nov. 2018. A new post doc in the lab: Shunichi KASHIDA. Welcome.
- April 2018. Publication in ACS Synthetic Biology
- Feb. 2018 Marc, post doc in the group, received funding from a Marie Sklodowska-Curie grant. Congratulations!
- December 2017. Good news: André just got an ERC consolidator grant entitled 'Metabolic soft matter with life-like porperties'. It will start in june 2018.
- Nov. 2017 Georg, post doc in the group, received funding from DFG (Deutsche Forschungsgemeinschaft). Congratulations!
- August 2017. A new post doc in the lab: Marc Van Der Hofstadt. Welcome.
- May 2017. Publication in Nature Chemistry
- March 2017. A new post doc in the lab : Georg Urtel. Welcome.
- Jan. 2017. Publication in Nature Nanotechnology
- Oct. 2016. A new PhD student in the lab: Anis Senoussi. Welcome.
- Sept. 2016. Adrian is now a Doctor... Congratulations!
- Master and undergradate internships are always available. Please, contact us.